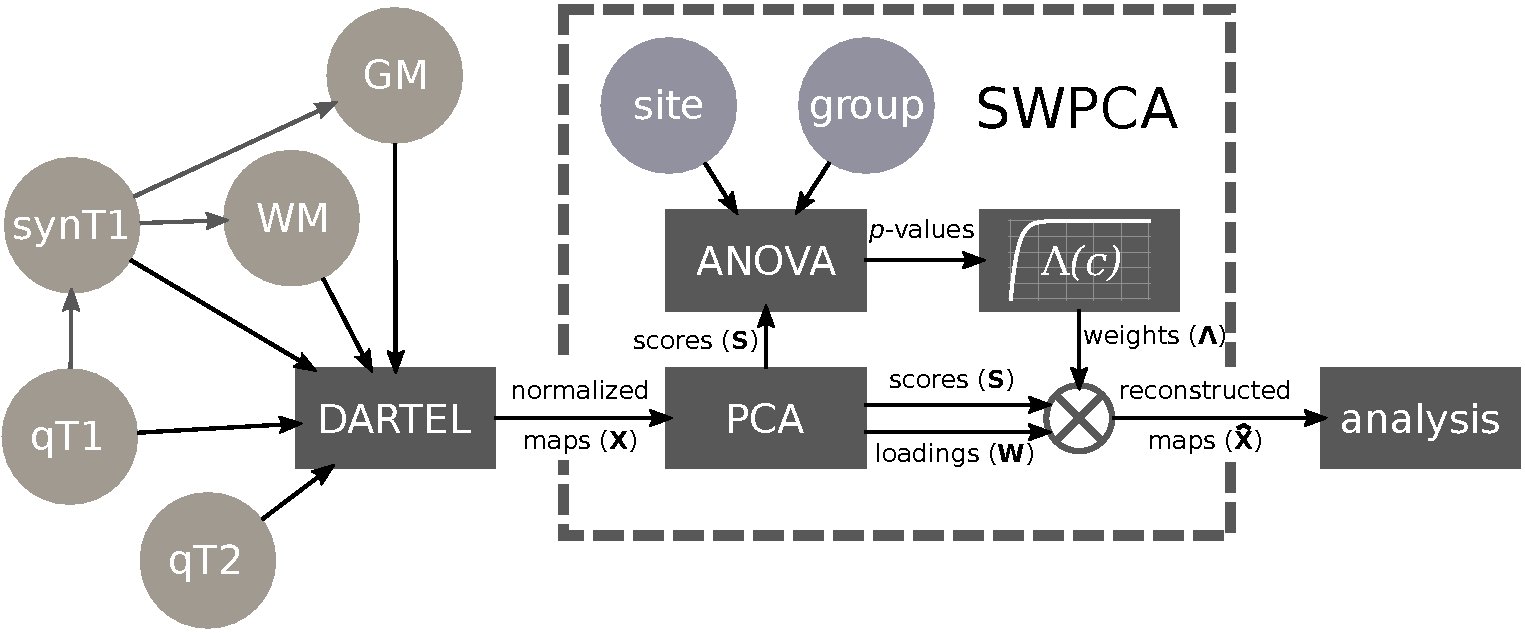
Significance Weighted Principal Component Analysis (SWPCA) is a technique developed to parse out the influence of a categorical variable that introduces variability in a certain dataset (Martinez-Murcia et al. 2017). This was originally intended to remove acquisition site variance in neuroimaging databases.
Download the code: Download
Use
To use the script to remove, navigate to the download dir, load the library (import swpca) into your environment and execute this command using the current dataset and acquisition site variables:
import swpca
dataset_rect, weights, A = swpca.swpca(dataset, site)It will return the rectified dataset, to be used in subsequent analysis.
Algorithm Pipeline and Context
The main use of the SWPCA algorithm is within the context of common neuroimaging analysis, such as Voxel Based Morphometry (VBM) or a classification analysis. It is a preprocessing step, and as such, it is used just after any other preprocessing steps (such as normalization, etc), and before any further analysis. It provides rectified maps with the influence of the categorical variables removed, therefore decreasing the occurrence of false positives.
References
Citation
@online{martinez-murcia2016,
author = {Martinez-Murcia, F.J.},
title = {Significance {Weighted} {Principal} {Component} {Analysis}
{(SWPCA)}},
date = {2016-10-24},
url = {https://pakitochus.github.io/fjmartinezmurcia.es/posts/2016-09-15-project-sbm/},
langid = {en}
}